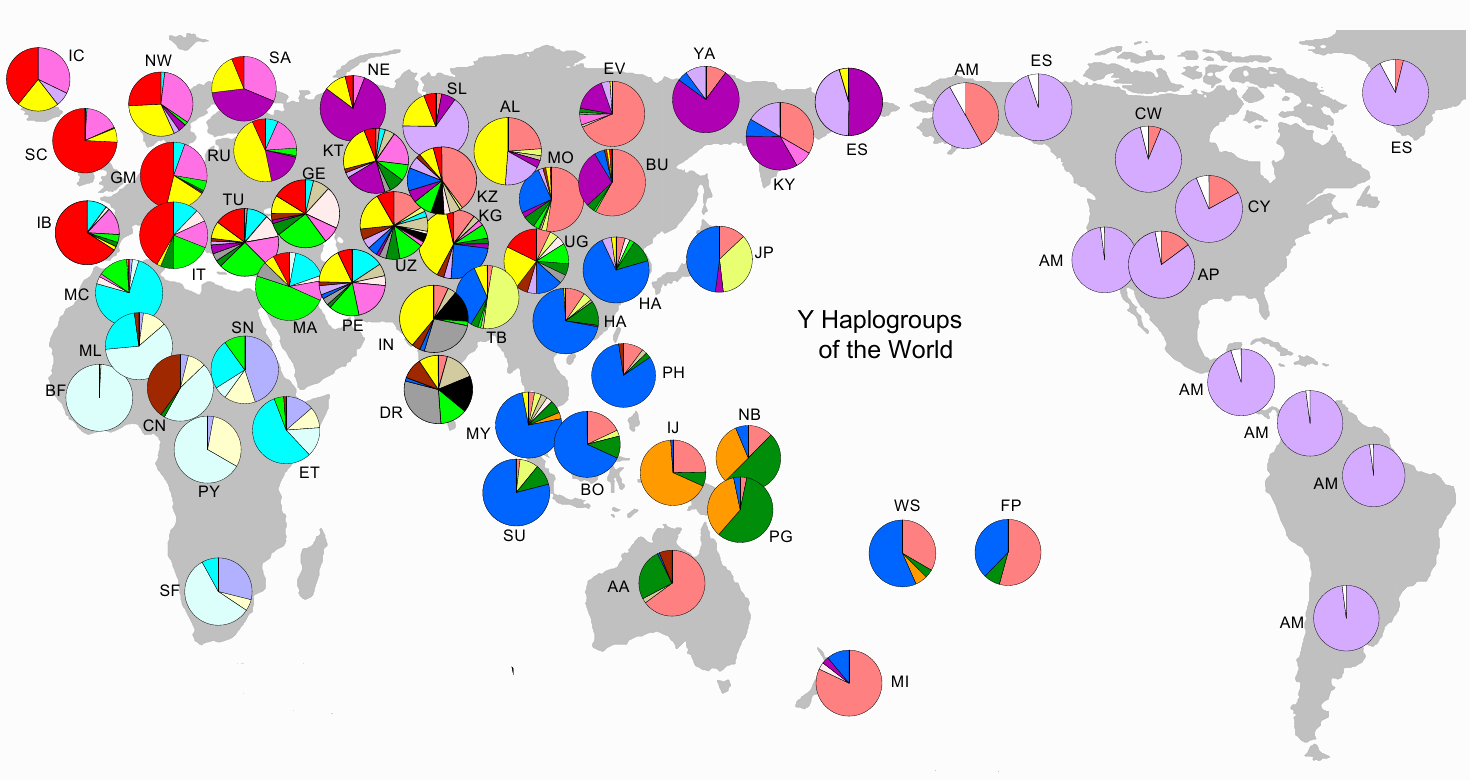
Y haplogroups
What follows is the first in a series of two essays, covering human haplogroups, their origins and current distributions. According to geneticists, no more than 90% of existing human mtDNA and yDNA haplogroups has been precisely identified. One theory is that the missing 10% was acquired through the archaic interbreeding between humans and at least two, non-human species. To me, this conjecture smacks of an argument from ignorance, i.e. we do not know where these haplogroups arose; therefore, we (sorta) know where they came from. But, the speculation remains fodder for thought.
- yDNA haplogroups
- clusters of non-recombinant DNA from the Y chromosome passed down the male line
yDNA haplogroups are used as genetic markers - in tracing the ancestry of male individuals to geographically distributed populations. Haplogroups are not known to be visible to selection; that is, they are traits, carried by individuals, which do not confer either survival or reproductive advantages. (Nor are they known to confer survival nor reproductive disadvantages.) Their frequencies are driven by genetic drift. All identified Y haplogroups are the results of down-stream mutations altering the original, human haplogroup (A) - now estimated to have arisen 140Kyr ago in one male, Adam (the most recent common male ancestor).
In the image below are striking clues, pointing towards drift. The pie-charts represent the relative frequency of a haplogroup (or haplogroups) in a given region. As examples - in the Americas among male Amer-Indians, haplogroup Q1a3a1 (light purple) is the most common, and in sub-Saharan West Africa, E1b1a (light blue) is the most common Y haplogroup. The high frequency distributions of these haplogroups on both continents are expectable - in that the Americas and sub-Saharan Africa were (largely) reproductively isolated for tens of thousands of years from the rest of the world. See: World Haplogroups.
The remaining account will be a compact summary of Y haplogroups with my comments and extensions - where necessary.
A BT B CT CF C D E F G H IJ I J K L M N O P Q R S T
140Kyr in North East or South West Africa
current populations:
Namibia (San 66%), Khoisan 44%, Mbuti ("pygmy"), Namibia (Nama 64%), Sudan (Dinka, Shilluk and Nuba) and Ethiopian Jews
The sub-clades of A are:
A0
mutation: P305
current populations: Cameroon (Bakola "pygmy") and Algeria (Berbers)
A1
mutation: L985
A1a-M31
mutation: M31
current populations: Guinea-Bissau, Senegambia (Mandinka) and Mali (Dogon)
A1b1a1a-M6
mutation: M6
current populations: Khoisan and Nama
A1b1b2b-M13
mutation: M13
current populations: Sudan (Nuba and Hausa), Ethiopia (Amhara)
A1a-M32
mutation: M32
current populations: Eastern and Southern Africa
In Africa, A1a-M32 is found at high frequency in large populations, whose male members carry A, but outside of Africa - in Turkey, Egypt, Palestine, Jordan, Oman, Yemen (Jews) and Sardinia, A1a-M32 shows up at low frequencies in small (localized) populations.
Complete Khoisan and Bantu genomes from southern Africa
70-80Kyr in North West or central West Africa
mutation:
M42
BT has not been found in any current population. No male has been shown to carry BT (BT*).
note
In Y haplogroups, paragroups are represented by an asterisk " * ", placed after the main haplogroup nomenclature. Paragroups contain the mutations which define the parent haplogroup, but they do not have any further (known) unique markers. Without these unique markers, they do not form truly independent sub-clades.
60-65Kyr in Central Africa
mutation:
M60
current populations:
B is localized among the Baka and Mbuti peoples of the tropical forests of West-Central Africa and the Hadza of Tanzania. 2.3% of African-American males carry B.
B is the second oldest and a very diverse Y haplogroup, but it is scattered widely and thinly in Africa, suggesting that the carriers of B were displaced by later (5Kyr) flows of people and events. A competing hypothesis runs that the sub-Saharan population dwindled (to ~2K persons at 35Kyr) and that there were few remaining carriers of B around to have been displaced by even much later migrations of (Bantu) people.
Some of the sub-clades of B are:
B1
mutation: M236
current population: southern Cameroon (Bamileke 4%)
B1a
mutation: M146
current population: Burkina Faso (Mossi 2%)
B2
mutation: M182
current populations: Congo (Mbuti), southern Cameroon (Bakola), Namibia (Dama) and Central African Republic (Biaka "pygmy")
B2a
mutation: M150
current populations: Congo (Mbuti 8%), Cameroon (Tupuri 11%), Mali (Dogon 6%) and Kenya (Kikuyu and Kamba 2%)
B2a1
mutation: M218
current population: northern Cameroon
B2a1a
mutation: M109
current populations: Cameroon, Central African Republic, Tanzania, Kenya, Ethiopia, South Africa, Zimbabwe, Sudan, Egypt (2%), Southern Iran (3%), African-Americans (1.5%), Pakistan and India
B2b
mutation: M112
current populations: Central African Republic (Baka "pygmy" 67%), Tanzania (Hadza 51%), Congo (Mbuti 43%), Namibia (San 31%)
B2b4
mutation: P7
current populations: Central African Republic (Baka 67% and Biaka 45%) and Congo (Mbuti 21%)
B2b4b
mutation: MSY2.1
current populations: Central African Republic (Biaka 20%)
68.5Kyr in East Africa
mutation:
M168
CT is often referred to as the "Eurasian Adam" - the most recent common ancestor of all non-African males. This hypothetical male is conjectured to have existed in Africa - immediately prior to the exodus of Anatomically Modern Humans from Africa. CT is the considered the common ancestral lineage of most men living today - though no male has been shown to carry CT (CT*).
The mutations M168, P9.1 and M294 have been found in all males tested - with the exception of those exclusively carrying A and B (sub-Saharan) haplogroups.
note
Paragroup (CT*) contains the mutations which define the parent haplogroup (M168, P9.1 and M294), but it does not have any further (known) unique markers.
55-65Kyr in Southwest Asia
mutation:
P143
current populations:
No male has been shown to carry CF (CF*). However, CF is the hypothetical ancestor of haplogroups C and F.
50-60Kyr in the Middle East or South Asia
mutation:
M130
current populations:
Northern Eurasia, Eastern Eurasia, Oceania and the Americas
C appeared shortly after humans expanded from Africa, and it may have originated and diversified in India or along the coasts of South Asia. After the African exodus, C (as with D below) was spread as a "Great Coastal Migration" from Arabia to Southeast Asia then northward into East Asia.
C*
mutation: M130
current populations: India, Sri Lanka and South East Asia
The sub-clades of C are:
C1
mutation: M8
current population: the rarest lineage of C. Found in Japan.
C2
mutation: M38
current populations: New Guinea, Melanesia and Polynesia
C2a1
mutation: P33
current populations: high frequency in Polynesia
C3
mutation: M217
current populations: Northern Asia, Japan (Ainu 15%), the Americas and Eastern and Central Europe
C3 was probably spread to Europe by the Huns in the Middle ages.
C3a2
mutation: P39
current populations: Amer-Indians (Na-Dene, Algonquian and Siouan-speaking populations)
C3a3
mutation: M48
current populations: Siberia, Mongolia and Central Asia
C4
mutation: M347
current population: aboriginal Australians
C5
mutation: M356
current population: India
65Kyr in Africa or Asia
mutation:
M145
current populations:
Africa and East Asia (very rare)
65-70Kyr in Asia
mutation:
M174
current populations:
Central Asia, Southeast Asia and Japan
D like C is correlated with the "Great Coastal Migration" from Arabia to Southeast Asia then northward into East Asia. Found today at high frequency in Tibet, Japan and the Andaman Islands.
The sub-clades of D are:
D1
origin: Asia
mutation: M15
current populations: Central Asia, East Asia, and Southeast Asia
Tibet (12.5%) and China (Quang 23%)
D2
origin: Asia
mutation: M55
current populations: Ainu, Japanese (35%) and Ryukyuans
D3
origin: Asia
mutation: P99
current populations: China (Pumi and Naxi) and Tibet
50-55Kyr in East Africa or the Near East
mutation:
M96
current populations:
sub-Saharan, North and North East Africa, the Near East and Europe
Originating in North East Africa or the Middle East, E was later introduced to West Africa - where it spread (5Kyr ago) to Central, Southern and South Eastern Africa with the Bantu migrations, swamping (or displacing) populations, whose members carried A and B.
The sub-clades of E are:
E1
origin: 45-55Kyr in Africa
mutations: L504, L511, P147
E1a
origin: 40-45Kyr
mutations: L633, M33, M132
E1b
origin: 45Kyr
mutation: P177
E1b1
mutation: P178
current populations: sub-Saharan Africa, North Africa, the Near East and Europe
E1b1a
mutation: V38
current populations: E1b1a is the most common Y haplogroup in sub-Saharan Africa, reaching a frequency of ~99% in Central West Africa.
E1b1b
mutation: M215
current populations: About 14Kyr, E1b1b spread throughout North and North East Africa, the Near East and later Europe. E1b1b is the third most frequent Y haplogroup in Europe.
E2
mutation: M75
current populations: E2 is found in East, Southern, Central and West Africa. The highest frequencies are among Bantu males of Kenya and South Africa.
45Kyr in North Africa, the Middle East or South Asia
mutation:
M89
current populations:
North Africa, the Near East, Europe and South Asia
F is frequently referred to as "the second-wave out of Africa". F is the parent of all Y haplogroups G through T, and it contains more than 90% of the world's non-sub-Saharan male population. Some male populations carrying F later migrated back to North Africa. For a discussion of the bi-directional gene flow between North Africa and the Near East - see: The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations.
The sub-clades of F are:
F1
mutation: P91
F2
mutation: M427
F3
mutation: L279
F4
mutation: M481
15-35Kyr in Near East or Southern Asia
mutation:
M201
current populations:
Iran, the Caucasus (~60% of Ossetian males), ~10% of Jewish males, Turkey and Pakistan
G was one of the "F-scale" haplogroups injected into the (R1b and I) populations of Old Europe, during the Neolithic expansion of peoples from the Near East - about 10Kyr ago.
The sub-clades of G are:
G1
origin: 5Kyr in Iran
mutation: M285
current populations: Iran, Turkey, Kazakhstan and the southern and Northern Caucasus
G1 is relatively rare in Europe.
G2
origin: possibly 3Kyr in Anatolia
mutation: P287
current populations: Caucasus, Southwest and Southern Asia
G2 is more common than G1.
G2a1
mutation: L149.1
G2a1a
mutation: L293
current populations: Caucasus, Eastern Europe and Ashkenazi Jews
G2a1b
mutation: L223
current populations: Southwest and southern Asia, Corsica and Sardinia
Oetzi, the Iceman preserved for over 5Kyr in the icy Italian Alps, belongs to G2a1b.
G2a1c1
mutation: M406
current populations: Turkey (5%), Greece (5%), Iraq (Kurds), Italy, Spain, Netherlands and Switzerland
G2a1c1a1
mutation:
current populations: Europe and Turkey (Armenia)
Haplogroup G Project
25-45Kyr in India, Iran or the Middle East
mutation:
M69
current populations:
Europe (Romani), India and Sri Lanka
H1
mutation: M52
current populations: India (Dravidians 33%), Sri Lanka (Sinhalese) and Nepal
H1a
mutation: M82
current populations: Europe (Romani people), India and Cambodia
35-40Kyr Southwest Asia
mutation:
M429
IJ is a hypothetical haplogroup, considered to have given rise to I and J. Some speculate that when Cro-Magnon males entered Europe (~40Kyr), they carried IJ. Under this theory - after it picked up a mutation, the dominant halplogroup in the male population of Europe (prior to the Last Glacial Maximum, LGM) ~25Kyr was I1.
25-30Kyr in Europe or the Middle East
mutation:
M170
current populations:
I is carried by the descendants of men who are believed to have arrived in Europe from the Middle East 20-25Kyr ago. They were associated with the Gravettian culture (22-28Kyr). I is the second most common Y haplogroup in North West Europe after R1b. 25% of males in Europe: the Balkans, Germany, Scandinavia and North Western Europe carry I. (Bosnia and Herzegovina 65%, Norway 40%, Denmark, 39%, Germany 24% and England 20%)
However, a competing theory runs that the parent of I (IJ) was the oldest Y haplogroup to appear in Europe - and that it (not R1b) was carried by the descendants of Cro-Magnon - at about 25Kyr.
The sub-clades of I are:
I1
origin: 15-25Kyr in Europe
mutation: M253
current populations: found in 35% of the Scandinavian population (Southern Norway, South Western Sweden and Denmark), Iceland and Northwestern Europe
I1 is associated with the Viking conquest of Britian.
I2
origin: 15Kyr in Poland or south eastern Europe
mutation: M438
current populations: Bosnia and Herzegovina, Croatia, Serbia, Sardinia, Spain (Basques), Denmark, Germany and Sweden
Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe
30Kyr in Southwest Asia
mutation:
M304
current populations:
Arabia, the Near East, Southern Europe, Central Asia, South Asia, North Africa and the Horn of Africa
The distribution of haplogroups J, R1b and T among the ancient (pre-Western colonial) populations of Africa is closely correlated with the language distribution of the Afro-Asiatic superfamily.
The sub-clades of J are:
J1
origin: 15-24Kyr in Western Asia
mutation: M267
current populations: Southwest Asia, North Africa and Ethiopia
J2
origin: 18.5Kyr in Turkey or Fertile Cresent
current populations: Turkey, the Levant, Mesopotamia, the South Caucasus, Iran, Central Asia and South Asia
J2 spread into the Mediterranean area with the influx of agricultural peoples from the Near East during the early Neolithic (~10Kyr). 29% of Sephardic Jews and 23% of Ashkenazi Jews carry J2.
47Kyr in South Western or Central Asia
mutation:
M9
current populations:
Asia, Europe and the Americas
25-30Kyr in Iran or Southern Central Asia
mutation:
M20
current populations:
India (Dravidian upper and middle castes), Pakistan, the Near East and Europe
L may have been (with the exception of J2) the original Y haplogroup of the creators of the Indus Valley Civilization.
32-47Kyr in Southeast Asia
mutation:
P256
current populations:
Indonesia, Melanesia, Micronesia and Polynesia
In Western New Guinea, M is the most frequent male haplogroup.
15-20Kyr in Southeast Asia
mutation:
M231
current populations:
Siberia, Eurasia and Europe (Finland 60%, Latvia and Lithuania 40%, Russia 20%)
The sub-clade of N is:
N1
mutation: LLY22g.1_1
N1c1
mutation: M46
current population: Siberia and northern Europe
N1c2a
mutation: M128
current populations: Kazakhstan, Korea and China
35Kyr in Siberia or Central Asia
mutation:
M175
current populations:
80-90% of all men in East and Southeast Asia carry O.
The sub-clades of 0 are:
O1
mutation: MSY2.2
current populations: Malaysia, Vietnam, Indonesia and southern China
O2
mutation: L463
current populations: Japan and Korea
O3
mutation: M122
current populations: China
27-41Kyr in Central Asia or Southern Siberia
mutation:
M48
P is the parent haplogroup of R and Q. It contains the patrilineal ancestors of most Europeans and most Amer-Indians.
15-20k in Siberia
Humans colonized Siberia by ~45Kyr. It's striking that 10-20Kyr after the African exodus, some of its descendants appear to have made a bee-line straight to (and successfully inhabited) Siberia - one of the coldest regions on the planet. Routinely - during Siberian winters, temperatures drop to -60F. Haplogroup Q (and possibly O and P) arose in Siberia.
mutation:
M242
current populations:
Amer-Indians and North Eurasians
Q is the most common haplogroup among Amer-Indian males.
The sub-clade of Q is:
Q1
Q1a3a1
origin: Beringia 10-15Kyr
mutation: M3
current population: Q1a3a1 is almost exclusively associated with the Amer-Indian population. Though found in Siberia at low frequencies, it may have been the result of ancient back-flow from North America - before Amer-Indians became reproductively isolated from the rest of the world.
THE PEOPLING OF THE NEW WORLD: Perspectives from Molecular Anthropology
20-35Kyr in Central or South Asia
mutation:
M207
current populations:
Europe, Central and South Asia, the Middle East and Africa
The sub-clades of R are:
R1
origin: 12-25Kyr in Central or South Asia
mutation: M173
current populations: Europe, Western Asia, Africa, Siberia and the Americas
R1 is relatively common among male Amer-Indians - in North Eastern Canada and the US, triggering speculation that R1 was brought to the Americas recently during the time of the European Conquest.
R1 is believed to have existed long before the end of last Ice Age. It has been associated with the Aurignacian culture (40-25Kyr). Archeological evidence supports the view that the Aurignacian culture arrived from Anatolia during the Upper Paleolithic (rather than earlier theories which tied this culture to the Iranian plateau). The Aurignacian culture and Cro-Magnon, the first modern humans to enter Europe 40-35Kyr, are linked. However, the contention that Cro-Magnon males carried R1 has been challenged. Any link, connecting R1 to the Aurignacian culture, is weak - as some estimates suggest that R1 arose only 18.5Kyr ago.
R1a
origin: 18.5Kyr in Asia, South Asia, Central Asia, Middle East or Eastern Europe
mutation: M420
current populations: Its distribution is associated with the re-settlement of Eurasia following the LGM - 18-22Kyr.
R1a1a
origin: 18.5Kyr in the Eurasian Steppes
mutation: M17
current populations: R1a1a, common in Europe, is associated with the expansion of the Kurgan people who spread Indo-European languages to Central Asia, India, Sri Lanka, Central, Northern and Eastern Europe. The Kurgans were pastoral nomads, who rode the horse and chariot, shot a compound bow, smelted bronze and worshipped the sky god. They conquered (or co-opted) many cultures, notably Greece and the Indus Valley civilization. They also invaded Babylon, establishing the 500 year long Kassite dynasty.
R1b
origin: 18.5Kyr in Western Asia
mutation: M343
current populations: R1b is the most common Y haplogroup in the male, Western European population. The present-day population of Western Europe, carrying R1b, is believed to have descended from a "refugium" in the Iberian Peninsula (Portugal and Spain) during the LGM - where the R1b1b2 haplogroup achieved a "genetic homogeneity". After the ice sheets receded in Europe, these R1b carrying males (in part) re-colonized Europe. However, see a contrary discussion regarding R1b here. It's - further - speculated that in Old Europe the dominant Paleolithic (pre-LGM) Y haplogroup was IJ (not R1b).
R1b1
mutation: P25
R1b1b2
origin: 18.5Kyr(?) in Central Asia or South Central Siberia
mutation: P25 and M269
current populations: Most of the present-day European males carrying the M343 (R1b) marker also have the P25 and M269 markers, which define the R1b1b2 as a subclade.
R1b1*
mutation: P25-derived
current populations: Northern Cameroon
R1b1* represents archaic gene flow from Eurasia into sub-Saharan Africa (~22Kyr). Modern-day populations of Northern Cameroon speak Chadic languages, which are classified as an ancient branch of the Afro-Asiatic superfamily of languages. The extinct language of the Ancient Egyptians belonged to this superfamily.
A Back Migration from Asia to Sub-Saharan Africa Is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes
R2
origin: 25Kyr in South Central Asia
mutation: M124
current populations: India, Pakistan and Sri Lanka
The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans
28-41Kyr in Southeast Asia (New Guinea)
mutation:
M230
current populations:
New Guinea (~50%), Indonesia and Melanesia
The sub-clade of S is:
S1
mutation: M254
19-34Kyr in Western Asia
mutation:
M184
current populations:
India, Egypt, Oman, Tanzania, Ethiopia and Morocco
T is found at low frequencies in Europe and the Middle East. Thomas Jefferson, the 3rd President of the US, carried T.
The sub-clade of T is:
T1
mutation: M193